HLA-DRB1 Impact on Survival of Induced Leukemic Cells Under ‘Allergic’ Conditions
| Received 25 Nov, 2024 |
Accepted 15 Jan, 2025 |
Published 18 Jan, 2025 |
Background and Objective: Leukaemia is a hematological malignancy of which etiology is still unclear. Like most cancers, many are associated with a genetic defect such as chromosomal translocation. Recent epidemiology studies revealed an inverse relationship between acute lymphoblastic leukemia and allergic rhinitis (AR), a chronic inflammatory condition. This study investigates the impact of HLA-DRB1 alleles on the survival of induced leukemic cells under allergic conditions. Materials and Methods: In this study, a Dermatophagoides pteronyssinus (Der p 1)-sensitized AR subject typed as HLA-DRB1*13 (susceptible allele in AR) and a non-allergic subject with HLA-DRB1*10 (protective allele in AR) of Malay ethnic group were recruited. Peripheral Blood Mononuclear Cells (PBMC) were transfected with the oncogenic MLL/MLLT1 fusion gene to induce leukemia cells. The induced leukaemia-like B cells were co-cultured for five days with Der p1 stimulated PBMC from the same individual. Survival of the B leukemic-like cells was analyzed by flow cytometry. Three experiments were performed, non-parametric statistical analyses: Mann-Whitney for two-group comparisons was used with significance level, p<0.05 Results: Induced leukemic-like cells were CD34lowCD38+CD45+MLL+CD19+. This population was significantly reduced in HLA-DRB1*13 samples compared to HLA-DRB1*10 (1.07+0.12% vs 3.53+0.61%, p = 0.05). Significantly decreased regulatory T cells (Tregs) in HLA-DRB1*13 compared to HLA-DRB1*10 (4.93+0.12% vs 6.20+0.17%, p = 0.05) implied increased inflammatory condition. Conclusion: Thus, HLA-DRB1 alleles may be factors that influence the inverse relationship between allergic rhinitis and acute lymphoblastic leukemia. Further studies with a larger sample size are required to confirm the results.
| Copyright © 2025 Idris et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Leukaemia is a hematological malignancy or blood cancer detected in blood and bone marrow associated with a rise in immature and abnormal white blood cells. An estimated approximately 2.5% of new cancer leukemia are diagnosed every year1. Leukaemia may be categorized as acute or chronic and of lymphoid or myeloid lineage. Acute lymphoblastic leukemia (ALL) is most common among children making up 75% of patients. Peak incidence occurs between ages one to four years2. ALL is characterized by immature B or T cell blasts in the bone marrow, lymph nodes, thymus or spleen. The majority are of the B-cell lineage. Chronic leukemias, such as chronic lymphocytic leukemia (CLL), affect mature cells and exhibit varying incidence rates based on age, gender, and race, with about 10% linked to a family history3. The CLL typically occurs in individuals aged 60-80, with 10% under 55. Peripheral blood smear reveals small mature lymphocytes with numerous smudge cells. Lymphoma is a primary malignant neoplasm of lymphoid tissues divided as Hodgkin’s lymphoma and Non-Hodgkin’s lymphoma (NHLs)4.
In 2020, a total of 83,087 new cases of Hodgkin lymphoma were reported and there was an increasing trend, especially among females, younger populations, and Asian countries5. While non-Hodgkin’s lymphoma has an incidence of nearly 3% of cancers diagnosed worldwide6. The NHLs are classified into B and T cells and diagnosed by immunophenotyping markers such as CD45, CD20, CD3, CD24. Classical Hodgkin’s lymphoma is classified as nodular sclerosis, mixed cellularity, lymphocyte rich, and lymphocyte depleted. Markers such as CD21, CD23, CD3, CD4, CD57, BCL6, EMA are used to diagnose the four variants7.
Knowledge of etiology of leukemia is limited. Predisposing factors include inherited genetic susceptibility and environmental risk2. Other than environment and cancer-causing viruses, aberrant genetics has the potential to influence leukemia development8. Behavioral factors and lifestyle such as smoking, obesity, and physical inactivity may increase an individual’s risk of developing cancer.
Evidence is beginning to show that an inverse association exists between childhood leukemia with allergic rhinitis (AR)9. In the USA, examining 1842 ALL cases and 1986 matched controls, suggests that allergic disorders may be associated with reduced risk of childhood ALL10. The United Kingdom Childhood Cancer Study (UKCCS) reported that, an approximately 20% reduction in risk for ALL and common ALL (c-ALL) was observed11.
Allergic rhinitis (AR) is a mucosal inflammation of the nasal area in patients due to inhalation and immune reaction to allergens. A common airborne allergen is house dust mite (Dermatophagoides sp.). Clinical symptoms are sneezing, rhinorrhoea, nasal obstruction, and pruritus12. Early response in AR, a type 1 hypersensitivity, is mediated by allergen-specific immunoglobulin E (IgE) activated via T cells specific to allergens presented on HLA (human leukocyte antigen). Activation of T cells polarises to T helper 2 (Th2) subtype and production of cytokines including IL-4, IL-5, IL-6, and IL-13. These cytokines recruit other arms of the immune response such as eosinophils, basophils, and neutrophils, and result in the deregulation of the epithelial cell barrier integrity which creates an allergic inflammation loop that maintains inflammation upon continued exposure to inflammatory antigens13. This inflammatory condition is suspected to exert a bystander effect that prevents the survival of pre-leukemia cells and the reduction of ALL among AR children.
The inherited highly polymorphic HLA genes predispose to various inflammatory diseases. In certain diseases the association is so strong that the HLA gene is used as a diagnostic marker e.g., ankylosing spondylitis (HLA-B27), celiac disease (HLA-DQ2 and DQ8), and narcolepsy (HLA-DQB*06:02). The main source of polymorphism is contributed by the six major genes, HLA class-I (A, B, and C) and HLA Class-II (DP, DQ, DR). To date, the IPD-IMGT/HLA Database has reported more than 3,700 alleles for HLA-DRB1 alone.
In ALL, the HLA-DRB1*04 allele appears to be a female-specific susceptibility factor for the acquisition of childhood ALL, while the HLA-DRB1*11 allele may have prognostic significance in childhood ALL14. HLA-DRB1*03, *16 were identified as risk alleles while *07 and *12 were reported to be protective among Malays15. Allergic rhinitis also demonstrated association with HLA particularly HLA-DQB1*06:01:01 and HLA-DRB1*08:03:02 were considered as risk alleles for house dust mite (HDM)-sensitized AR, while HLA-DQB1*05 was protective in Chinese Han.
The association between ALL and AR, however, is unconfirmed and remains controversial. Given that HLA genes confer susceptibility and protection effects on diseases, there is a possibility that HLA may be a determining factor in the development of the disease. This study aimed to determine in vitro survival of induced leukemic cells in the allergic environment of patients selected for specific HLA alleles.
MATERIALS AND METHODS
Study area: Participants were recruited in the Otorhinolaryngology Clinic, Hospital Serdang, Malaysia. This study was carried out from January, 2019 to March, 2022.
Samples: It was hypothesized, the inverse association of acute lymphoblastic leukemia (ALL) and allergic rhinitis (AR) patients may be influenced by susceptible/protective HLA-DRB1 alleles. A non-allergic control with HLA-DRB1*10 (haplotype DRB1*10, DRB1*15) and an allergic rhinitis patient with HLA-DRB1*13 (haplotype DRB1*13, DRB1*15) were recruited. Screening was done using protocol from Bunce et al.16. Whole blood was collected and Peripheral Blood Mononuclear Cells (PBMC) were isolated by the density gradient centrifugation method using Ficoll-Paque PLUS (GE Health Care, Sweden).
Induced leukemia cells: Leukemia-like cells were generated by transfection of PBMC with the acute lymphoblastic leukemia-associated fusion gene, MLL/MLLT1 using lipofectamine assay. At least 200,000 PBMC cells were seeded in 1 mL of growth medium RPMI 1640 (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and antibiotics consisting 2 μg/mL Gentamicin (Gibco), 1.25 μg/mL Amphotericin B (Gibco) and 100 U/mL penicillin-streptomycin (Gibco) in a 24 well plate and incubated overnight in 5% CO2, at 37°C. The transfection reagent was prepared immediately before transfection. Two μg of plasmid DNA MLL/MLLT1 was diluted in 100 μL of serum-free Roswell Park Memorial Institute (RPMI). The transfection reagent (Lipofectamine 2000 and LTX reagent (Thermofisher, USA) was briefly vortexed and 5 μL of each reagent was added to the diluted DNA. This was immediately mixed by pipetting or vortexing the DNA mixture and then incubated for 5 min at room temperature. Following that, 100 μL of the transfection reagent/DNA mixture was added dropwise to each well-containing cells in the RPMI medium. The plate was gently rocked to achieve an even distribution of the complexes immediately after adding the transfection reagent and the plate was incubated at 37°C in 5% CO2. For short-term transient transfection, cells were grown in a standard medium (RPMI 1640) for 10 days.
Co-culture experiment: The ‘allergic environment’ consisted of the in vitro culture of PBMC from the selected individuals with specific allergens. PBMC (1 million cells) was stimulated with 5 μg/mL LoToxTMNatural Der p1 (Invitrogen) extract (INDOOR Biotechnologies, UK) antigen for four days in 1 mL RPMI 1640, 10% fetal bovine serum (FBS) (Gibco, US) and antibiotics consisting of 2 μg/mL Gentamicin (Gibco, US), 1.25 μg/mL Amphotericin B (Gibco, US) and 100 U/mL penicillin-streptomycin (Gibco, US). Cultures were kept at 37°C with 5% CO2. A non-stimulated control was included.
At the end of 4 days, 300 μL of induced leukemia cells were added to 300 μL of Der p1 stimulated PBMC and co-cultured for another 5 days at 5% CO2 at 37°C. Cells were then harvested.
Flow cytometry analysis: Induced cells were identified by intracellular expression of the KMT2A/ENL fusion protein (from transfected MLL/MLLT1 fusion gene) using the polyclonal antibody MLL/KMT2A (Novus, USA). Surface marker expression of B-lineage leukemia-associated markers was determined with monoclonal antibodies CD19 PE, CD34 PE Cy7, CD45 PerCy 5.5, and CD38 APC-H7 (BD Biosciences, USA). Surface and intracellular staining methods were done according to the standard protocols. For intracellular staining, cells were permeabilized with 1 mL of 1×Perm wash (BD, Biosciences, USA) and fixed with one mL of Fix buffer/Paraformaldehyde (PFA) (BD, Biosciences, USA). Cells were then acquired on a flow cytometer (FACSCanto) and analyzed with the FACSDiva Software (BD Biosciences, US). A total of 5,000 events were collected in the lymphocyte region of co-cultured cells. Stimulated PBMC as the ‘allergic environment’ was monitored for levels of regulatory T cells (Tregs) as a marker of the inflammatory condition. The Tregs marker cocktail was CD4/CD25/CD127 (BD Biosciences, USA).
Statistical analysis: Data, expressed as Mean±SD, underwent non-parametric statistical analyses: Mann-Whitney for two-group comparisons. One-sided analysis was performed using SPSS software and a significance level of p<0.005.
RESULTS
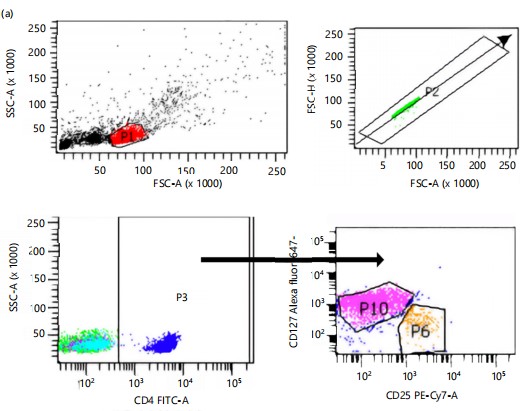
Flow cytometry gating strategy for regulatory T (Tregs) and T effector cells are shown in Fig. 1. *Figure 1 illustrates the gating strategy and results of regulatory and effector T cell analysis. In Fig 1a, lymphocyte-like cells (P1) were identified by gating on low side scatter, followed by the selection of singlet cells (P2). Helper T cells/CD4-positive cells were gated on P3, and regulatory T cells (Tregs) were identified as CD4+/CD25+/CD127– (P6), while T effector cells were identified as CD4+/CD25+/CD127+. Figure 1(b-c) show that regulatory T cells (CD4+CD25+CD127–) were significantly higher in HLA-DRB110 individuals compared to HLA-DRB113, both before (6.20±0.17% vs 4.93±0.12%) and after (7.30±2.07 vs 4.97±0.06) co-culture, respectively. Conversely, Fig. 1(d-e) reveal no significant differences in T effector cells (CD4+CD25+CD127+) between HLA-DRB110 and HLA-DRB113 individuals, either before (61.47±2.20% vs 70.20±4.51%) or after (64.40±4.27% vs 67.40±5.97%) co-culture.
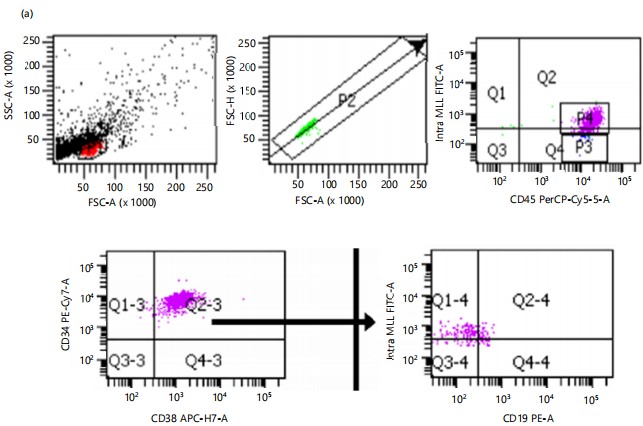
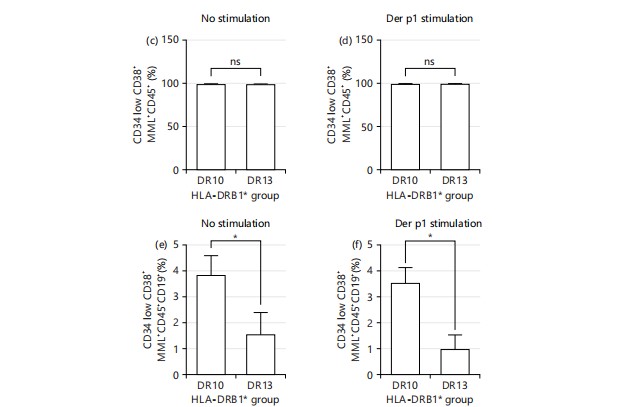
Figure 2 a-b show dot plot flow cytometric analysis of the transfected population (CD34lowCD38+MLL+CD45+) and abnormal B cell populations (CD34lowCD38+MLL+CD45+CD19+) for HLA-DRB110 and HLA-DRB113, respectively, with gating on low side scatter to identify lymphocyte-like cells (P1) followed by selection for singlet cells (P2). In both conditions, statistical analyses (Fig. 2 c-d) revealed no significant differences in the percentages of CD34lowCD38+MLL+CD45+ cells between HLA-DRB110 and HLA-DRB113, both without and with Der p1 stimulant. However, Fig. 2e shows that the abnormal B cell population (CD34lowCD38+MLL+CD45+CD19+) was significantly reduced in the HLA-DRB113 individual under unstimulated conditions compared to the HLA-DRB110 individual (3.50+1.11 vs 1.57+0.84). Similarly, Fig. 2f illustrates a significant reduction in the abnormal B cell population in the HLA-DRB113 individual under Der p1 stimulant conditions compared to HLA-DRB110 (3.53+0.61% vs 1.07+0.12%).
Transfection of PBMC from the HLA-DRB1*10 (non-allergic) and HLA-DRB1*13 (allergic rhinitis) individuals with the MLL/MLLT1 fusion gene resulted in CD19+ B cells with abnormal growth. The transfected cells were co-cultured with prior Der p1 stimulated PBMC from the same individual. Non-stimulated PBMC was used as a control. Figure 2a-b show the flow cytometric analysis of the transfected population (CD34lowCD38+MLL+CD45+) and abnormal B cell populations (CD34lowCD38+MLL+CD45+CD19+) before and after Der p1 stimulation.
DISCUSSION
In this study, Peripheral Blood Mononuclear Cells (PBMC) samples were used as a source of B cells. The PBMC isolated from peripheral blood samples consists of B, T, monocytes as well as Tregs. The PBMC is important as a tool in both research and diagnosis and is used in short-term and long-term cultures17,18. Between myeloid cells, T cells, and B cells, B cells are more difficult to transfect19. Isolated B cells require special culture media with growth hormones likely to increase the cost of materials. Prolonged culture also exposes B cells to contamination and cell death. On the other hand, the short-term culture of PBMC without stimulants is commonly used. The T cells and monocytes already in the cultures create a microenvironment suitable for immune cell growth. Multiple cytokines (interleukins, IL-7, IL-4, IL-6, IL-10, and interferons, IFN-α, IFN-β, IFN-γ) produced and memory T cells upon activation play important roles in the development, survival, differentiation, and/or proliferation of B cells20. Thus, it was hypothesized T cells and monocytes may provide growth factors to maintain B cells in vitro.
House-dust mite (HDM) is an important indoor source of allergic reactions including allergic rhinitis (AR)21. In this study, Der p1 allergen was used to stimulate PBMC. The Der p1 is a cysteine proteinase derived
|
|
Tregs a subpopulation of T cells with immune suppressor function form 5-10% of CD4 positive mature T cells. Tregs function as suppressors and are important to restrain and shut down inflammatory and fatal autoimmune responses throughout the lifespan of an organism23. In cancers, including hematological malignancies, it may exert major immunosuppressive activity which plays a critical role in tumor cell growth, proliferation, and survival24. In allergic rhinitis, an inflammatory condition, there is evidence of a reduction in the Tregs population25.
There is evidence that Tregs numbers are decreased in AR26-28. Suppressor cytokine, IL-10 was lower in nasal samples from AR compared to the healthy control group29. More importantly, an immunotherapy trial to compare immunological responses to specific subcutaneous immunotherapy (SCIT) and sublingual immunotherapy (SLIT) towards CD4+CD25+Foxp3+ show upregulation of these cells indicated good clinical improvement30. Immunopathology of allergy is reportedly an imbalance between type 2 immune response and Tregs31. Thus, it was not surprising that the (CD4+CD125+CD127–) Tregs in the allergy rhinitis sample (HLA-DRB1*13) were found to be significantly lower than the non-allergic sample (HLA-DRB1*10) further supporting earlier observations. There was no difference in the T effector (CD4+CD125+CD127+) population, suggesting this may not be the critical population in effector function in allergic rhinitis which may lie with other effector cells such as the IgE-producing B cells.
The diagnosis of leukemia makes use of monoclonal antibody markers to identify cell lineage and determine stages of differentiation. Normal cells with bright CD34, nuclear TdT, low CD10, and CD45 suggest an early progenitor stage. Subsequently, B-cells are CD34– and TdT-negative, maintain low CD10, and increase CD20 and CD45 expression. In the third stage, B cells express high CD20 with dim CD10. On the other hand, CD58, CD38, CD24, CD19, and CD22 are expressed in all three stages. An increase in CD10 and CD58 and a decrease in CD38 and CD45 are observed in pediatric B-ALL patients32. The CD34 and CD38 expression can be used as biomarkers for the prognosis of B-cell ALL. Expression of CD34 is low and high expression of CD38, is associated with good/favorable prognosis32. Characterization in peripheral blood, bone marrow, and secondary lymphoid tissues is marked by the accumulation of small mature B cells with CD5+ in CLL33,34. Other leukemia markers to detect CLL are a high expression of CD5, CD19, and CD23. Prognostic markers for CLL are CD38, CD25, CD56, and CD117. Several of the markers that were used here identified the growth of an abnormal population of B cells that were MLL+ but maintained an original level of expressions for CD34, CD38, CD45, and CD19 of mature cells.
The fusion gene t (11;19) (q23;p13.3) or MLL/MLLT1 (MLL/ENL) has been used frequently to create leukemia cells. Primary human hematopoietic stem and progenitor cells (HSPC) from bone marrow, cord blood, or even induced pluripotent stem cells were sources of early cells used to replicate acute leukemia cell models) this chromosomal abnormality tended to generate B-cell acute lymphoblastic leukaemia35. Here, PBMC was attempted as the cell source. As observed, transfection with the leukemia MLL/MLLT1 fusion gene did not induce an acute leukemia phenotype but generated an abnormal B cell leukemia-like population. Nevertheless, combining cloning with a leukemia-associated oncogene and samples with an identified genetic background such as an HLA-DR allele was able to generate cell models that were more specific as tools for further studies and understanding leukemia better.
Co-culture experiments replicate the leukemia cells in an allergic microenvironment. The co-cultures compared the genetic microenvironment of HLA-DRB1*10, the allele associated with protection against allergic rhinitis and risk for acute lymphoblastic leukemia allele, and HLA-DRB1*13 allele, susceptible to allergic rhinitis and lower incidence of acute lymphoblastic leukemia compared to healthy individuals. A significantly reduced percentage of MLL+ B cells in the HLA-DRB1*13 co-cultures supported the hypothesis that the inflammatory condition in the allergic samples did not support the survival of leukemic cells. This finding is novel and has not been reported before. The significantly reduced Tregs in the HLA-DRB1*13 samples were another factor that indicated a higher inflammatory condition here. Increased levels of Tregs are observed in various cancers including ALL35. Future studies should explore the underlying mechanisms by which HLA-DRB1 alleles affect T cell regulation in allergic conditions and their impact on leukemia survival. Expanding the sample size and incorporating longitudinal data would provide more robust insights. Limitations of this study include its preliminary nature, small sample size, and the need for further validation in diverse populations to ensure the generalizability of the findings.
CONCLUSION
Significantly reduced Tregs cells (CD4+CD25+CD127–) in the HLA-DRB1*13/allergic rhinitis PBMC cultures compared to HLA-DRB1*10/non-allergic PBMC implied an increased inflammatory microenvironment. A significantly reduced percentage of MLL+CD19+ leukemic-like cells in the HLA-DRB1*13 cultures supported the hypothesis that this increased inflammation condition prevented the survival of the leukemic cells.
Thus, HLA-DRB1 alleles may be factors that influence the inverse relationship between allergic rhinitis and acute lymphoblastic leukemia. Further studies should explore the molecular mechanisms underlying HLA-DRB1's role in leukemic cell survival under various immune-modulatory conditions to develop targeted therapeutic strategies.
SIGNIFICANCE STATEMENT
This study suggests that HLA-DRB1*13/allergic rhinitis PBMC cultures exhibit reduced Treg cells and an increased inflammatory environment, potentially inhibiting leukemic cell survival. The findings propose that HLA-DRB1 alleles may influence the inverse relationship between allergic rhinitis and acute lymphoblastic leukemia. Further research with larger sample sizes is needed to confirm these results.
REFERENCES
- Malard, F. and M. Mohty, 2020. Acute lymphoblastic leukaemia. Lancet, 395: 1146-1162.
- Dong, Y., O. Shi, Q. Zeng, X. Lu, W. Wang, Y. Li and Q. Wang, 2020. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp. Hematol. Oncol., 9.
- Sheikhpour, R., F. Pourhosseini, H. Neamatzadeh and R. Karimi, 2017. Immunophenotype evaluation of non-Hodgkin’s lymphomas. Med. J. Islam Republic Iran, 31: 804-807.
- Huang, J., W.S. Pang, V. Lok, L. Zhang and D.E. Lucero-Prisno III et al., 2022. Incidence, mortality, risk factors, and trends for Hodgkin lymphoma: A global data analysis. J. Hematol. Oncol., 15.
- Thandra, K.C., A. Barsouk, K. Saginala, S.A. Padala, A. Barsouk and P. Rawla, 2021. Epidemiology of non-Hodgkin’s lymphoma. Med. Sci., 9.
- Grewal, R.K., M. Chetty, E.A. Abayomi, C. Tomuleasa and J.R. Fromm, 2019. Use of flow cytometry in the phenotypic diagnosis of Hodgkin's lymphoma. Cytometry Part B: Clin. Cytometry, 96: 116-127.
- Milan, T., H. Canaj, C. Villeneuve, A. Ghosh, F. Barabé, S. Cellot and B.T. Wilhelm, 2019. Pediatric leukemia: Moving toward more accurate models. Exp. Hematol., 74: 1-12.
- Yari, F., M. Sobhani, F. Sabaghi, M. Zaman-Vaziri, N. Bagheri and A. Talebian, 2008. Frequencies of HLA-DRB1 in Iranian normal population and in patients with acute lymphoblastic leukemia. Arch. Med. Res., 39: 205-208.
- Wen, W., X.O. Shu, M.S. Linet, J.P. Neglia, J.D. Potter, M.E. Trigg and L.L. Robison, 2000. Allergic disorders and the risk of childhood acute lymphoblastic leukemia (United States). Cancer Causes Control, 11: 303-307.
- Hughes, A.M., T. Lightfoot, J. Simpson, P. Ansell and P.A. McKinney et al., 2007. Allergy and risk of childhood leukaemia: Results from the UKCCS. Int. J. Cancer, 121: 819-824.
- Meng, Y., C. Wang and L. Zhang, 2019. Recent developments and highlights in allergic rhinitis. Allergy, 74: 2320-2328.
- Small, P., P.K. Keith and H. Kim, 2018. Allergic rhinitis. Allergy Asthma Clin. Immunol., 14.
- El Ansary, M.M., L.A. Mohammed, T.H. Hassan, A. Baraka and A.A. Ahmed, 2015. Human leukocyte antigen-DRB1 polymorphism in childhood acute lymphoblastic leukemia. Mol. Clin. Oncol., 3: 425-429.
- Hassan, N., S.Z. Idris, K.M. Chang, R. Osman, H.M. Ibrahim, J.S. Dhaliwal and M. Abdullah, 2024. High variability in HLA-DRB1*03, a predisposing allele in acute lymphoblastic leukemia. Iran. J. Blood Cancer, 16: 24-33.
- Prasad, V., M. Abdullah, F. Nordin and S.T. Subha, 2022. Prevalence, causes and treatments of allergic rhinitis in Malaysia: A literature review. Egypt. J. Otolaryngology, 38.
- Bunce, M., C.M. O'Neill, M.C.N.M. Barnardo, P. Krausa, M.J. Browning, P.J. Morris and K.I. Welsh, 1995. Phototyping: Comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens, 46: 355-367.
- Stabel, J.R. and T.L.T. Wherry, 2023. Comparison of methods to isolate peripheral blood mononuclear cells from cattle blood. J. Immunol. Methods, 512.
- Bueno, C., J.L. Sardina, B. di Stefano, D. Romero-Moya and A. Muñoz-López et al., 2016. Reprogramming human B cells into induced pluripotent stem cells and its enhancement by C/EBPα. Leukemia, 30: 674-682.
- Vazquez, M.I., J. Catalan-Dibene and A. Zlotnik, 2015. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine, 74: 318-326.
- Sidenius, K.E., T.E. Hallas, L.K. Poulsen and H. Mosbech, 2001. Allergen cross-reactivity between house-dust mites and other invertebrates. Allergy, 56: 723-733.
- Sun, R., X.Y. Tang and Y. Yang, 2016. Immune imbalance of regulatory T/type 2 helper cells in the pathogenesis of allergic rhinitis in children. J. Laryngology Otology, 130: 89-94.
- Plitas, G. and A.Y. Rudensky, 2016. Regulatory T cells: Differentiation and function. Cancer Immunol. Res., 4: 721-725.
- D’Arena, G., C. Vitale, M. Coscia, A. Festa and N.M.D. di Minno et al., 2017. Regulatory T cells and their prognostic relevance in hematologic malignancies. J. Immunol. Res., 2017.
- Palmer, C., J.K. Mulligan, S.E. Smith and C. Atkinson, 2017. The role of regulatory T cells in the regulation of upper airway inflammation. Am. J. Rhinology Allergy, 31: 345-351.
- Xu, G., Z. Mou, H. Jiang, L. Cheng and J. Shi et al., 2007. A possible role of CD4+CD25+ T cells as well as transcription factor Foxp3 in the dysregulation of allergic rhinitis. Laryngoscope, 117: 876-880.
- Lee, J.H., H.H. Yu, L.C. Wang, Y.H. Yang, Y.T. Lin and B.L. Chiang, 2007. The levels of CD4+CD25+ regulatory T cells in paediatric patients with allergic rhinitis and bronchial asthma. Clin. Exp. Immunol., 148: 53-63.
- Shaoqing, Y., C. Yinjian, Y. Zhiqiang, Z. Ruxin, C. Na and G. Rongming, 2018. The levels of CD4+CD25+ regulatory T cells in patients with allergic rhinitis. Allergologie Select, 2: 144-150.
- Erkan, K., M.K. Bozkurt, H. Artaç, H. Özdemir, A. Ünlü, E.N. Korucu and Ç. Elsürer, 2020. The role of regulatory T cells in allergic rhinitis and their correlation with IL-10, IL-17 and neopterin levels in serum and nasal lavage fluid. Eur. Arch. Oto-Rhino-Laryngology, 277: 1109-1114.
- Xian, M., M. Feng, Y. Dong, N. Wei, Q. Su and J. Li, 2020. Changes in CD4+CD25+FoxP3+ regulatory T cells and serum cytokines in sublingual and subcutaneous immunotherapy in allergic rhinitis with or without asthma. Int. Arch. Allergy Immunol., 181: 71-80.
- Boonpiyathad, T., Z.C. Sözener, M. Akdis and C.A. Akdis, 2020. The role of Treg cell subsets in allergic disease. Asian Pac. J. Allergy Immunol., 38: 139-149.
- Wood, B.L., 2015. Flow cytometry in the diagnosis and monitoring of acute leukemia in children. J. Hematopathol., 8: 191-199.
- Jiang, Z., D. Wu, S. Lin and P. Li, 2016. CD34 and CD38 are prognostic biomarkers for acute B lymphoblastic leukemia. Biomarker Res., 4.
- Haq, H., Nasir Uddin, S.A. Khan and S. Ghaffar, 2020. Prognostic markers in chronic lymphocytic leukaemia-A flow cytometric analysis. Pak. J. Med. Sci., 36: 338-343.
- Buechele, C., E.H. Breese, D. Schneidawind, C.H. Lin and J. Jeong et al., 2015. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood, 126: 1683-1694.
- Idris, S.Z., N. Hassan, L.J. Lee, S.M. Noor and R. Osman et al., 2016. Increased regulatory T cells in acute lymphoblastic leukaemia patients. Hematology, 21: 206-212.
How to Cite this paper?
APA-7 Style
Idris,
S.Z., Prasad,
V., Amin,
A.M., Subha,
S.T., Mei,
L., Nordin,
F., Abdullah,
M. (2025). HLA-DRB1 Impact on Survival of Induced Leukemic Cells Under ‘Allergic’ Conditions. Singapore Journal of Scientific Research, 15(1), 1-10. https://doi.org/10.3923/sjsr.2025.01.10
ACS Style
Idris,
S.Z.; Prasad,
V.; Amin,
A.M.; Subha,
S.T.; Mei,
L.; Nordin,
F.; Abdullah,
M. HLA-DRB1 Impact on Survival of Induced Leukemic Cells Under ‘Allergic’ Conditions. Singapore J. Sci. Res 2025, 15, 1-10. https://doi.org/10.3923/sjsr.2025.01.10
AMA Style
Idris
SZ, Prasad
V, Amin
AM, Subha
ST, Mei
L, Nordin
F, Abdullah
M. HLA-DRB1 Impact on Survival of Induced Leukemic Cells Under ‘Allergic’ Conditions. Singapore Journal of Scientific Research. 2025; 15(1): 1-10. https://doi.org/10.3923/sjsr.2025.01.10
Chicago/Turabian Style
Idris, Siti, Zuleha, Vivek Prasad, Amrina Mohd Amin, Sethu Thakachy Subha, Lai Mei, Fazlina Nordin, and Maha Abdullah.
2025. "HLA-DRB1 Impact on Survival of Induced Leukemic Cells Under ‘Allergic’ Conditions" Singapore Journal of Scientific Research 15, no. 1: 1-10. https://doi.org/10.3923/sjsr.2025.01.10

This work is licensed under a Creative Commons Attribution 4.0 International License.

